Hui Ji
Annealed Relaxation of Speculative Decoding for Faster Autoregressive Image Generation
Jan 14, 2026Abstract:Despite significant progress in autoregressive image generation, inference remains slow due to the sequential nature of AR models and the ambiguity of image tokens, even when using speculative decoding. Recent works attempt to address this with relaxed speculative decoding but lack theoretical grounding. In this paper, we establish the theoretical basis of relaxed SD and propose COOL-SD, an annealed relaxation of speculative decoding built on two key insights. The first analyzes the total variation (TV) distance between the target model and relaxed speculative decoding and yields an optimal resampling distribution that minimizes an upper bound of the distance. The second uses perturbation analysis to reveal an annealing behaviour in relaxed speculative decoding, motivating our annealed design. Together, these insights enable COOL-SD to generate images faster with comparable quality, or achieve better quality at similar latency. Experiments validate the effectiveness of COOL-SD, showing consistent improvements over prior methods in speed-quality trade-offs.
FigEx2: Visual-Conditioned Panel Detection and Captioning for Scientific Compound Figures
Jan 12, 2026Abstract:Scientific compound figures combine multiple labeled panels into a single image, but captions in real pipelines are often missing or only provide figure-level summaries, making panel-level understanding difficult. In this paper, we propose FigEx2, visual-conditioned framework that localizes panels and generates panel-wise captions directly from the compound figure. To mitigate the impact of diverse phrasing in open-ended captioning, we introduce a noise-aware gated fusion module that adaptively filters token-level features to stabilize the detection query space. Furthermore, we employ a staged optimization strategy combining supervised learning with reinforcement learning (RL), utilizing CLIP-based alignment and BERTScore-based semantic rewards to enforce strict multimodal consistency. To support high-quality supervision, we curate BioSci-Fig-Cap, a refined benchmark for panel-level grounding, alongside cross-disciplinary test suites in physics and chemistry. Experimental results demonstrate that FigEx2 achieves a superior 0.726 mAP@0.5:0.95 for detection and significantly outperforms Qwen3-VL-8B by 0.51 in METEOR and 0.24 in BERTScore. Notably, FigEx2 exhibits remarkable zero-shot transferability to out-of-distribution scientific domains without any fine-tuning.
Aligning Findings with Diagnosis: A Self-Consistent Reinforcement Learning Framework for Trustworthy Radiology Reporting
Jan 06, 2026Abstract:Multimodal Large Language Models (MLLMs) have shown strong potential for radiology report generation, yet their clinical translation is hindered by architectural heterogeneity and the prevalence of factual hallucinations. Standard supervised fine-tuning often fails to strictly align linguistic outputs with visual evidence, while existing reinforcement learning approaches struggle with either prohibitive computational costs or limited exploration. To address these challenges, we propose a comprehensive framework for self-consistent radiology report generation. First, we conduct a systematic evaluation to identify optimal vision encoder and LLM backbone configurations for medical imaging. Building on this foundation, we introduce a novel "Reason-then-Summarize" architecture optimized via Group Relative Policy Optimization (GRPO). This framework restructures generation into two distinct components: a think block for detailed findings and an answer block for structured disease labels. By utilizing a multi-dimensional composite reward function, we explicitly penalize logical discrepancies between the generated narrative and the final diagnosis. Extensive experiments on the MIMIC-CXR benchmark demonstrate that our method achieves state-of-the-art performance in clinical efficacy metrics and significantly reduces hallucinations compared to strong supervised baselines.
Reinforced Diffusion: Learning to Push the Limits of Anisotropic Diffusion for Image Denoising
Dec 30, 2025Abstract:Image denoising is an important problem in low-level vision and serves as a critical module for many image recovery tasks. Anisotropic diffusion is a wide family of image denoising approaches with promising performance. However, traditional anisotropic diffusion approaches use explicit diffusion operators which are not well adapted to complex image structures. As a result, their performance is limited compared to recent learning-based approaches. In this work, we describe a trainable anisotropic diffusion framework based on reinforcement learning. By modeling the denoising process as a series of naive diffusion actions with order learned by deep Q-learning, we propose an effective diffusion-based image denoiser. The diffusion actions selected by deep Q-learning at different iterations indeed composite a stochastic anisotropic diffusion process with strong adaptivity to different image structures, which enjoys improvement over the traditional ones. The proposed denoiser is applied to removing three types of often-seen noise. The experiments show that it outperforms existing diffusion-based methods and competes with the representative deep CNN-based methods.
Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge
May 05, 2025


Abstract:Accurate fetal brain tissue segmentation and biometric analysis are essential for studying brain development in utero. The FeTA Challenge 2024 advanced automated fetal brain MRI analysis by introducing biometry prediction as a new task alongside tissue segmentation. For the first time, our diverse multi-centric test set included data from a new low-field (0.55T) MRI dataset. Evaluation metrics were also expanded to include the topology-specific Euler characteristic difference (ED). Sixteen teams submitted segmentation methods, most of which performed consistently across both high- and low-field scans. However, longitudinal trends indicate that segmentation accuracy may be reaching a plateau, with results now approaching inter-rater variability. The ED metric uncovered topological differences that were missed by conventional metrics, while the low-field dataset achieved the highest segmentation scores, highlighting the potential of affordable imaging systems when paired with high-quality reconstruction. Seven teams participated in the biometry task, but most methods failed to outperform a simple baseline that predicted measurements based solely on gestational age, underscoring the challenge of extracting reliable biometric estimates from image data alone. Domain shift analysis identified image quality as the most significant factor affecting model generalization, with super-resolution pipelines also playing a substantial role. Other factors, such as gestational age, pathology, and acquisition site, had smaller, though still measurable, effects. Overall, FeTA 2024 offers a comprehensive benchmark for multi-class segmentation and biometry estimation in fetal brain MRI, underscoring the need for data-centric approaches, improved topological evaluation, and greater dataset diversity to enable clinically robust and generalizable AI tools.
Multimodal Trustworthy Semantic Communication for Audio-Visual Event Localization
Nov 04, 2024



Abstract:The exponential growth in wireless data traffic, driven by the proliferation of mobile devices and smart applications, poses significant challenges for modern communication systems. Ensuring the secure and reliable transmission of multimodal semantic information is increasingly critical, particularly for tasks like Audio-Visual Event (AVE) localization. This letter introduces MMTrustSC, a novel framework designed to address these challenges by enhancing the security and reliability of multimodal communication. MMTrustSC incorporates advanced semantic encoding techniques to safeguard data integrity and privacy. It features a two-level coding scheme that combines error-correcting codes with conventional encoders to improve the accuracy and reliability of multimodal data transmission. Additionally, MMTrustSC employs hybrid encryption, integrating both asymmetric and symmetric encryption methods, to secure semantic information and ensure its confidentiality and integrity across potentially hostile networks. Simulation results validate MMTrustSC's effectiveness, demonstrating substantial improvements in data transmission accuracy and reliability for AVE localization tasks. This framework represents a significant advancement in managing intermodal information complementarity and mitigating physical noise, thus enhancing overall system performance.
RAG-RLRC-LaySum at BioLaySumm: Integrating Retrieval-Augmented Generation and Readability Control for Layman Summarization of Biomedical Texts
May 21, 2024Abstract:This paper introduces the RAG-RLRC-LaySum framework, designed to make complex biomedical research understandable to laymen through advanced Natural Language Processing (NLP) techniques. Our Retrieval Augmented Generation (RAG) solution, enhanced by a reranking method, utilizes multiple knowledge sources to ensure the precision and pertinence of lay summaries. Additionally, our Reinforcement Learning for Readability Control (RLRC) strategy improves readability, making scientific content comprehensible to non-specialists. Evaluations using the publicly accessible PLOS and eLife datasets show that our methods surpass Plain Gemini model, demonstrating a 20% increase in readability scores, a 15% improvement in ROUGE-2 relevance scores, and a 10% enhancement in factual accuracy. The RAG-RLRC-LaySum framework effectively democratizes scientific knowledge, enhancing public engagement with biomedical discoveries.
Multi-Center Fetal Brain Tissue Annotation (FeTA) Challenge 2022 Results
Feb 08, 2024



Abstract:Segmentation is a critical step in analyzing the developing human fetal brain. There have been vast improvements in automatic segmentation methods in the past several years, and the Fetal Brain Tissue Annotation (FeTA) Challenge 2021 helped to establish an excellent standard of fetal brain segmentation. However, FeTA 2021 was a single center study, and the generalizability of algorithms across different imaging centers remains unsolved, limiting real-world clinical applicability. The multi-center FeTA Challenge 2022 focuses on advancing the generalizability of fetal brain segmentation algorithms for magnetic resonance imaging (MRI). In FeTA 2022, the training dataset contained images and corresponding manually annotated multi-class labels from two imaging centers, and the testing data contained images from these two imaging centers as well as two additional unseen centers. The data from different centers varied in many aspects, including scanners used, imaging parameters, and fetal brain super-resolution algorithms applied. 16 teams participated in the challenge, and 17 algorithms were evaluated. Here, a detailed overview and analysis of the challenge results are provided, focusing on the generalizability of the submissions. Both in- and out of domain, the white matter and ventricles were segmented with the highest accuracy, while the most challenging structure remains the cerebral cortex due to anatomical complexity. The FeTA Challenge 2022 was able to successfully evaluate and advance generalizability of multi-class fetal brain tissue segmentation algorithms for MRI and it continues to benchmark new algorithms. The resulting new methods contribute to improving the analysis of brain development in utero.
Synthesis of realistic fetal MRI with conditional Generative Adversarial Networks
Sep 20, 2022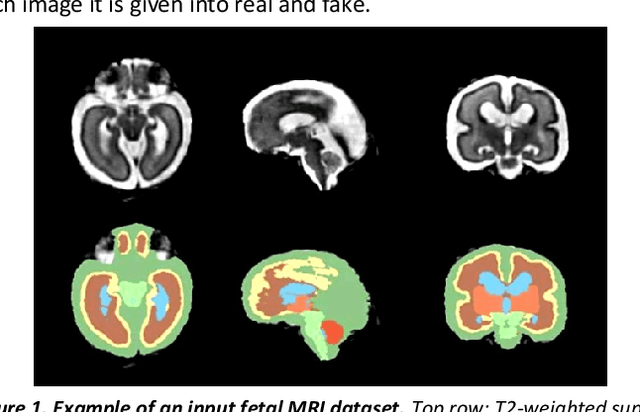
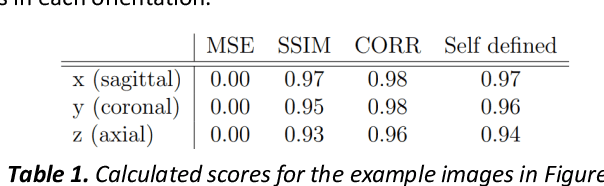
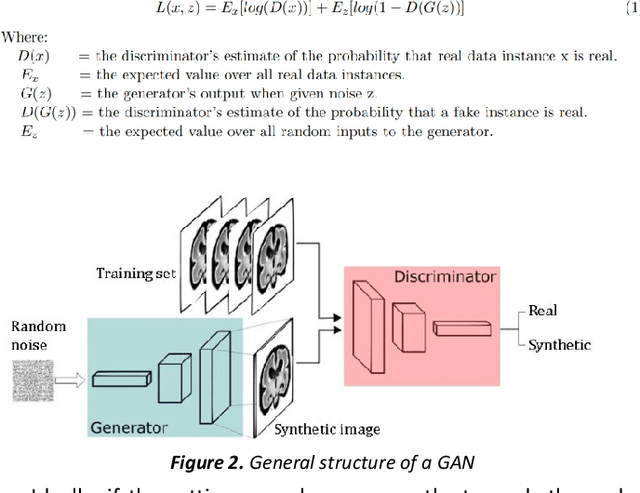
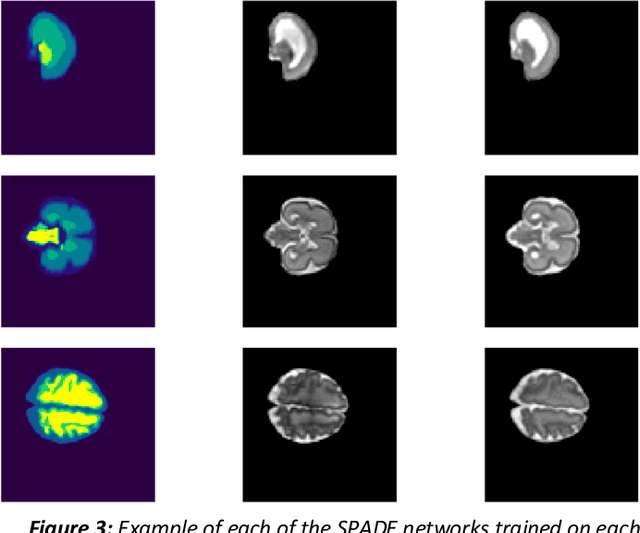
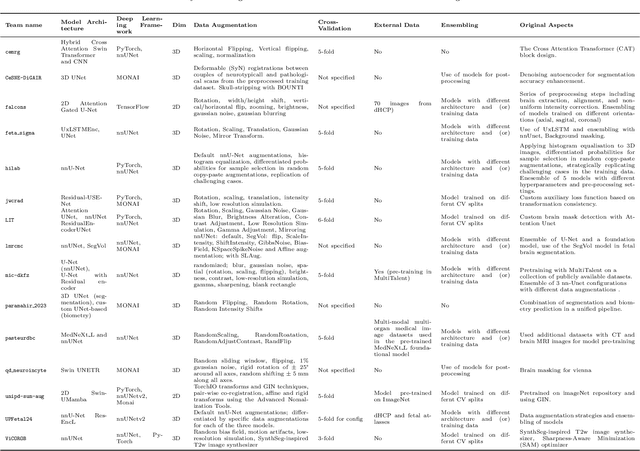
Abstract:Fetal brain magnetic resonance imaging serves as an emerging modality for prenatal counseling and diagnosis in disorders affecting the brain. Machine learning based segmentation plays an important role in the quantification of brain development. However, a limiting factor is the lack of sufficiently large, labeled training data. Our study explored the application of SPADE, a conditional general adversarial network (cGAN), which learns the mapping from the label to the image space. The input to the network was super-resolution T2-weighted cerebral MRI data of 120 fetuses (gestational age range: 20-35 weeks, normal and pathological), which were annotated for 7 different tissue categories. SPADE networks were trained on 256*256 2D slices of the reconstructed volumes (image and label pairs) in each orthogonal orientation. To combine the generated volumes from each orientation into one image, a simple mean of the outputs of the three networks was taken. Based on the label maps only, we synthesized highly realistic images. However, some finer details, like small vessels were not synthesized. A structural similarity index (SSIM) of 0.972+-0.016 and correlation coefficient of 0.974+-0.008 were achieved. To demonstrate the capacity of the cGAN to create new anatomical variants, we artificially dilated the ventricles in the segmentation map and created synthetic MRI of different degrees of fetal hydrocephalus. cGANs, such as the SPADE algorithm, allow the generation of hypothetically unseen scenarios and anatomical configurations in the label space, which data in turn can be utilized for training various machine learning algorithms. In the future, this algorithm would be used for generating large, synthetic datasets representing fetal brain development. These datasets would potentially improve the performance of currently available segmentation networks.
Dataset-free Deep learning Method for Low-Dose CT Image Reconstruction
May 01, 2022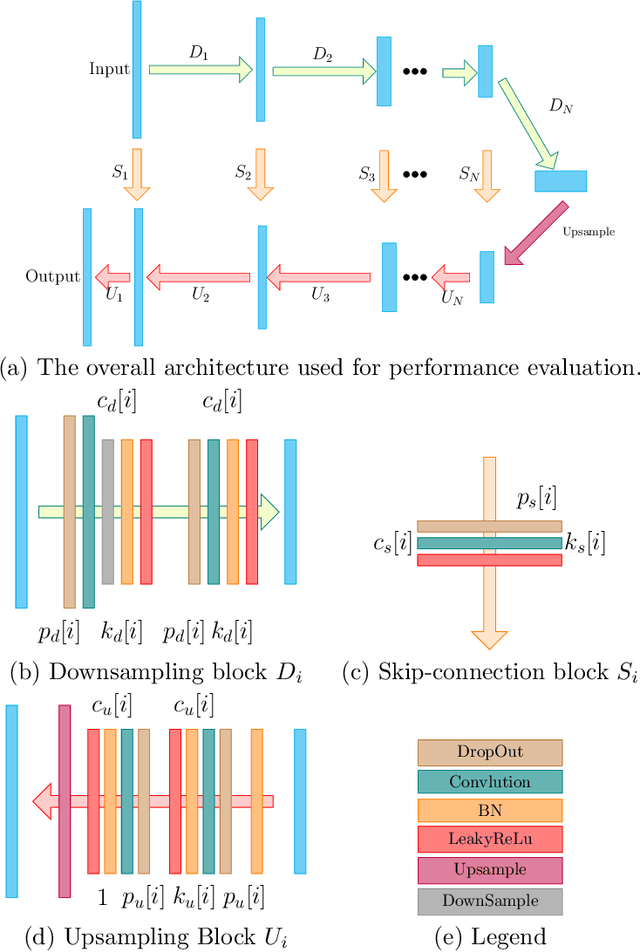
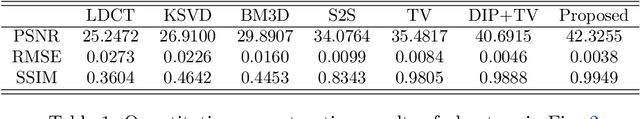
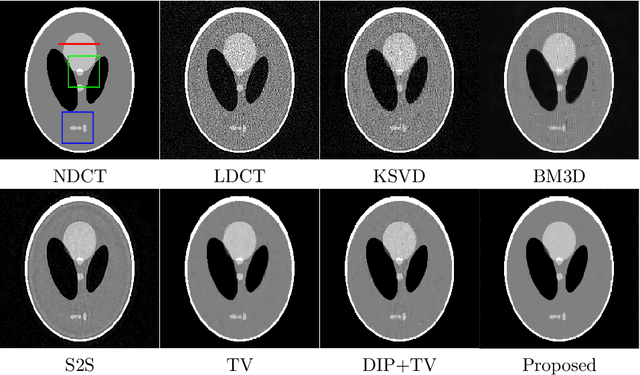
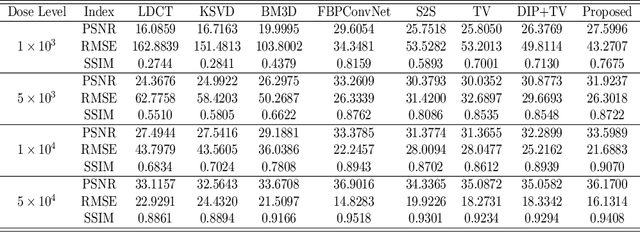
Abstract:Low-dose CT (LDCT) imaging attracted a considerable interest for the reduction of the object's exposure to X-ray radiation. In recent years, supervised deep learning has been extensively studied for LDCT image reconstruction, which trains a network over a dataset containing many pairs of normal-dose and low-dose images. However, the challenge on collecting many such pairs in the clinical setup limits the application of such supervised-learning-based methods for LDCT image reconstruction in practice. Aiming at addressing the challenges raised by the collection of training dataset, this paper proposed a unsupervised deep learning method for LDCT image reconstruction, which does not require any external training data. The proposed method is built on a re-parametrization technique for Bayesian inference via deep network with random weights, combined with additional total variational (TV) regularization. The experiments show that the proposed method noticeably outperforms existing dataset-free image reconstruction methods on the test data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge